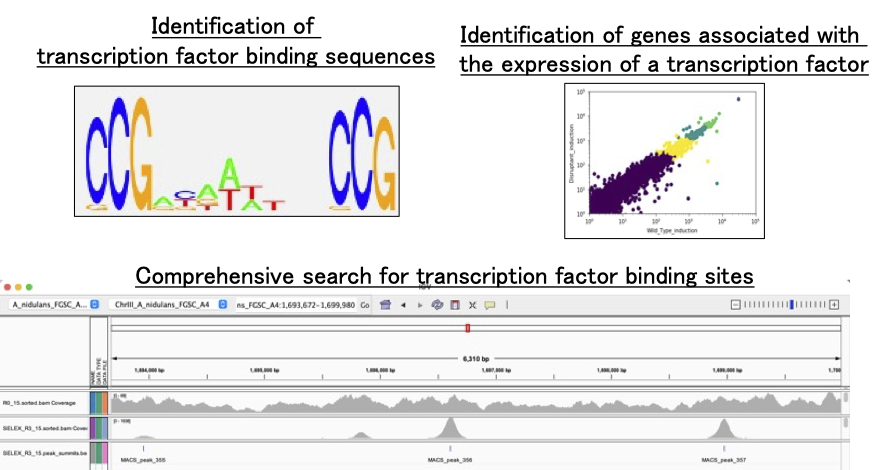
Transcriptional Regulatory Networks
We comprehensively map where DNA‑binding transcription factors engage in the genome and how they regulate downstream genes, using high‑throughput bioinformatics to decode transcriptional regulatory mechanisms.

We comprehensively map where DNA‑binding transcription factors engage in the genome and how they regulate downstream genes, using high‑throughput bioinformatics to decode transcriptional regulatory mechanisms.

Leveraging AI‑predicted structures (e.g. AlphaFold2) and experimental data, we build models to explain protein functions through their dynamic 3D conformations.
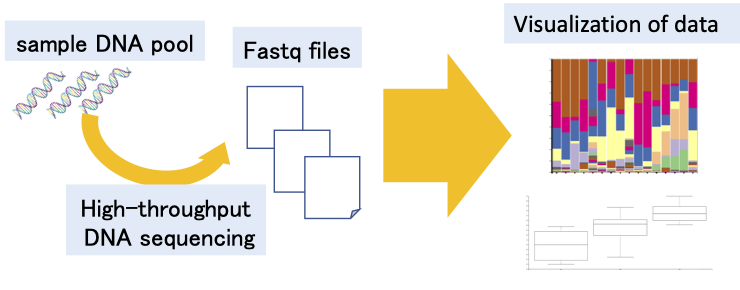
By analyzing environmental microbiomes with next‑gen sequencing, we uncover the complex relationships between plants and their surrounding microbial communities.
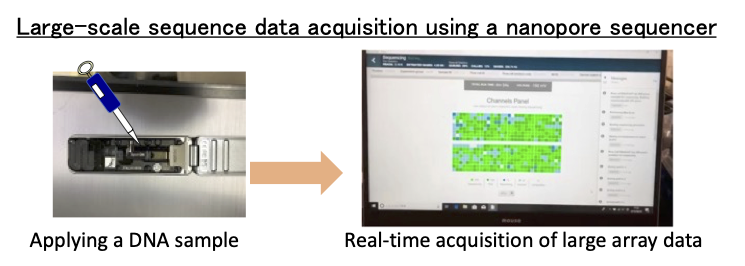
Utilizing long‑read sequencers, we perform large‑scale comparative genomics of environmental microbes to elucidate their ecological roles at the genome level.